Classification algorithms
Random Forest implementation using sklearn
source: Random Forests in Python
Note about the data: we will be using the famous iris data set which contains 4 variables measuring various parts of iris flowers (of 3 species) aas well as the species name.
Preliminar: load packages
In [1]:from sklearn.datasets import load_iris
from sklearn.ensemble import RandomForestClassifier
import pandas as pd
import numpy as np
Loading the data
In [2]:iris = load_iris()
df = pd.DataFrame(iris.data, columns=iris.feature_names)
df['species'] = pd.Categorical.from_codes(iris.target, iris.target_names)
df.head()
| sepal length (cm) | sepal width (cm) | petal length (cm) | petal width (cm) | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Creating the training and testing data
In [4]:df['is_train'] = np.random.uniform(0, 1, len(df)) <= .75
df.head()
| sepal length (cm) | sepal width (cm) | petal length (cm) | petal width (cm) | species | is_train | |
|---|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa | True |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa | True |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa | True |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa | True |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa | False |
train, test = df[df['is_train']==True], df[df['is_train']==False]
Preprocessing the data
In [6]:# Create a list of the feature column's names
features = df.columns[:4]
features
Index(['sepal length (cm)', 'sepal width (cm)', 'petal length (cm)',
'petal width (cm)'],
dtype='object')
# train['species'] contains the actual species names. Before we can use it,
# we need to convert each species name into a digit. So, in this case there
# are three species, which have been coded as 0, 1, or 2.
y, _ = pd.factorize(train['species'])
print(y)
Training the random forest classifier
In [9]:clf = RandomForestClassifier(n_jobs = 2 )
# Training the classifier
clf.fit(train[features], y)
RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',
max_depth=None, max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=10, n_jobs=2,
oob_score=False, random_state=None, verbose=0,
warm_start=False)
Applying to the data
In [10]:preds = iris.target_names[clf.predict(test[features])]
pd.crosstab(test['species'], preds, rownames=['actual'], colnames=['predicted'])
| predicted | setosa | versicolor | virginica |
|---|---|---|---|
| actual | |||
| setosa | 5 | 0 | 0 |
| versicolor | 0 | 11 | 0 |
| virginica | 0 | 2 | 12 |
View feature importance
In [12]:list(zip(train[features], clf.feature_importances_))
[('sepal length (cm)', 0.16992592921521485),
('sepal width (cm)', 0.019510194239802908),
('petal length (cm)', 0.18115102228639413),
('petal width (cm)', 0.62941285425858806)]
K- Means
Preliminar : load packages
In [13]:from sklearn.cluster import KMeans
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
Loading the data
In [14]:iris = load_iris()
# converting to a pandas DF for ease of use
x = pd.DataFrame(iris.data, columns=['Sepal Length', 'Sepal Width', 'Petal Length', 'Petal Width'])
y = pd.DataFrame(iris.target, columns=['Target'])
Exploratory visualization of the data
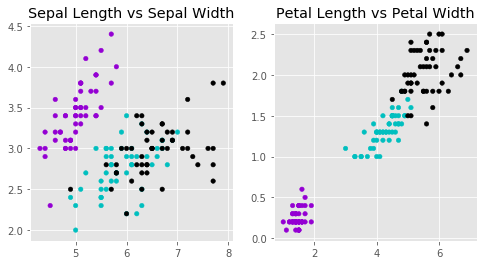
In [15]:# creating a plot of the data first
plt.style.use('ggplot')
fig, (ax1, ax2) = plt.subplots(1, 2, figsize = (8, 4), sharey =False)
colors = np.array(['darkviolet', 'c', 'black'])
#Draw a Scatter plot for Sepal Length vs Sepal Width
#nrows=1, ncols=2, plot_number=1
ax1.scatter(x['Sepal Length'], x['Sepal Width'], c=colors[y['Target']], s = 20)
ax1.set_title('Sepal Length vs Sepal Width')
ax2.scatter(x['Petal Length'], x['Petal Width'], c= colors[y['Target']], s = 20)
ax2.set_title('Petal Length vs Petal Width');
Create a model object consiting of 3 clusters
In [16]:model = KMeans(n_clusters = 3)
Apllying the model on the data
In [17]:model.fit(x)
KMeans(algorithm='auto', copy_x=True, init='k-means++', max_iter=300,
n_clusters=3, n_init=10, n_jobs=1, precompute_distances='auto',
random_state=None, tol=0.0001, verbose=0)
# model.labels_ contains the array of cluster ids
print (model.labels_)
Visualise the output of the model
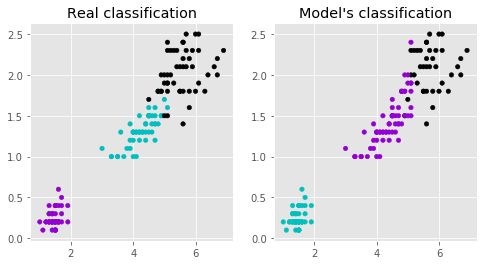
In [19]:fig, (ax1, ax2) = plt.subplots(1, 2, figsize = (8, 4), sharey =False)
predictedY = np.choose(model.labels_, [1, 0, 2]).astype(np.int64)
# Plot the classifications that we saw earlier between Petal Length and Petal Width
ax1.scatter(x['Petal Length'], x['Petal Width'], c=colors[y['Target']], s=20)
ax1.set_title('Real classification')
# Plot the classifications according to the model
ax2.scatter(x['Petal Length'], x['Petal Width'], c=colors[predictedY], s=20)
ax2.set_title("Model's classification");
import time
print('This notebook was last run on: ' + time.strftime('%d/%m/%y') + ' at: ' + time.strftime('%H:%M:%S'))
