BAD DAY 2: Generalized linear models
#for more examples see: http://plantecology.syr.edu/fridley/bio793/glm.html
setwd(getwd())Generalized linear models (GLMs) extend the linear modeling capability of R to scenarios that involve non-normal error distributions or heteroscedasticity.
All other classic assumptions (particularly independent observations) still apply. The idea here is that linear functions of the predictor variables are obtained by a link function. The data are then fit in this transformed scale (using an iterative routine based on least squares), but the expected variance is calculated on the original scale of the predictor variables.
Simple examples of link functions are $log(y)$ [which linearizes $exp(x)$], $sqrt(y) [x^2]$, and $1/y$. More particularly, GLMs work for the so-called ‘exponential’ family of error models: Poisson, binomial, Gamma, and normal.
Count (or count-like) response variables
dat = read.csv('./Data/treedata.csv') #choose the treedata.csv dataset
head(dat)
dim(dat)
dat2 = subset(dat,dat$species=="Tsuga canadensis")| plotID | date | plotsize | spcode | species | cover | utme | utmn | elev | tci | streamdist | disturb | beers |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATBN-01-0403 | 08-28-2001 | 1000 | ABIEFRA | Abies fraseri | 1 | 275736 | 3942439 | 1660 | 5.701460 | 490.9 | CORPLOG | 0.22442864 |
| ATBN-01-0532 | 07-24-2002 | 1000 | ABIEFRA | Abies fraseri | 8 | 302847 | 3942772 | 1712 | 3.823586 | 454.0 | VIRGIN | 0.83408785 |
| ATBN-01-0533 | 07-24-2002 | 1000 | ABIEFRA | Abies fraseri | 3 | 303037 | 3943039 | 1722 | 3.893762 | 453.4 | LT-SEL | 1.33325863 |
| ATBN-01-0536 | 07-25-2002 | 1000 | ABIEFRA | Abies fraseri | 3 | 273927 | 3935488 | 1754 | 3.145527 | 492.5 | SETTLE | 1.47124839 |
| ATBP-01-0001 | 05-11-1999 | 10000 | ABIEFRA | Abies fraseri | 8 | 273857 | 3937870 | 1945 | 5.682065 | 492.4 | VIRGIN | 1.64377141 |
| ATBP-01-0005 | 08-25-1999 | 10000 | ABIEFRA | Abies fraseri | 4 | 273876 | 3935462 | 1751 | 5.417182 | 545.9 | SETTLE | 0.00032873 |
- 8971
- 13
mean(dat2$cover)4.65951742627346
var(dat2$cover)4.471835471508
table(dat2$cover) 1 2 3 4 5 6 7 8 9 10
39 71 166 110 92 80 108 60 19 1
If these counts were distributed exactly from a Poisson process, what would they look like, assuming the same mean (and variance)?
bar1 = barplot(as.vector(table(dat2$cover)),names.arg=seq(1:10), col = 'lightblue2',
border = 'white')
points(bar1,dpois(seq(1,10),4.66)*sum(table(dat2$cover)),
cex=1,type="b", col = 'mediumpurple', lwd = 3, pch = 19)
#?dpois
box()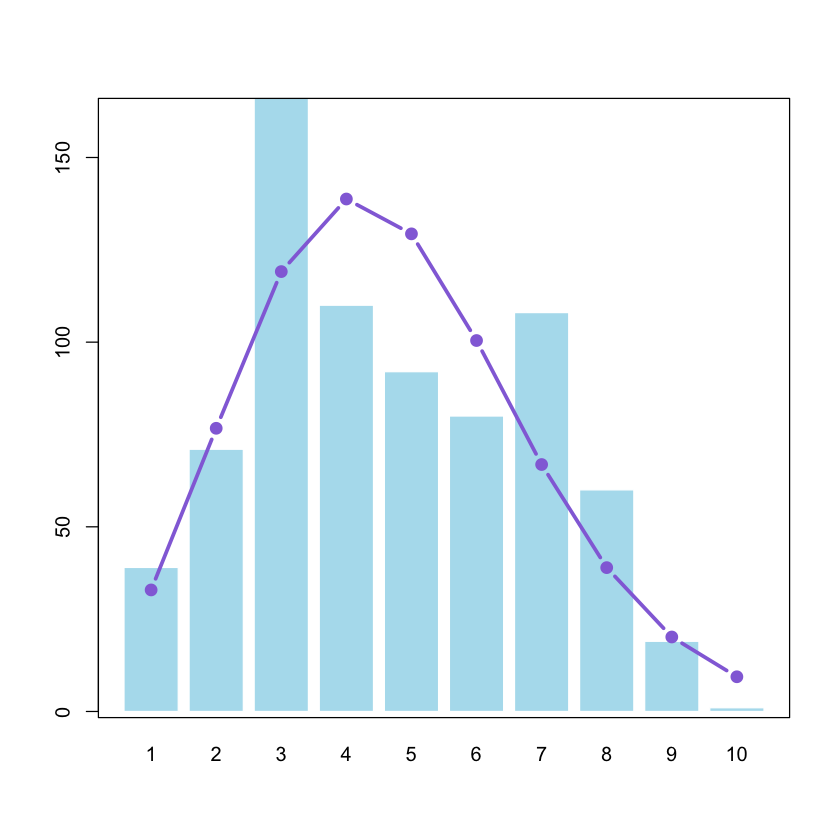
#?glmAs you can see, the data look Poisson-ish but they’re not perfect. (One reason you’ve probably already guessed: our data are bounded at 10, so variance should actually go down for the highest values.)
Now let’s fit a GLM to these data with just an intercept (overall mean):
glm1 = glm(cover~1,data=dat2,family=poisson)
summary(glm1)Call:
glm(formula = cover ~ 1, family = poisson, data = dat2)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.0594 -0.8229 -0.3132 1.0085 2.1430
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.53891 0.01696 90.73 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 749.25 on 745 degrees of freedom
Residual deviance: 749.25 on 745 degrees of freedom
AIC: 3212.2
Number of Fisher Scoring iterations: 4
Let’s add a continuous predictor variable like elevation to generate a simple Poisson regression. First we’ll graph it:
And then we’ll fit the new glm and test it against a model with only an intercept:
with(dat2,plot(elev,cover,main="Hemlock cover vs. elevation",
cex = 1.5, col = 'midnightblue', pch = 19))
Note the chi-squared test is typically recommended for models with ‘known deviance’ (Poisson and binomial). Here the model with elevation adds no explanatory power (fairly obvious from the graph), but we can still add the predicted trend line to our graph:
x = seq(0,1660)
#plot.new()
#lines(predict(glm2,list(elev=x),type="response"),lwd=2,col="orange")
#What is crucial here is the type argument to predict: "response" re-calculates the coefficients to be on the same scale as the original response variable, rather than the scale of the link function. Let's now try an ANOVA with Poisson error, using disturbance as our predictor:
glm3 = glm(cover~disturb, data = dat2, family = poisson)
summary(glm3)
anova(glm1, glm3, test = "Chisq")Call:
glm(formula = cover ~ disturb, family = poisson, data = dat2)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.1794 -0.7763 -0.1980 0.8523 2.2006
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.48367 0.03838 38.661 <2e-16 ***
disturbLT-SEL 0.03204 0.04685 0.684 0.494
disturbSETTLE 0.08957 0.05485 1.633 0.103
disturbVIRGIN 0.12184 0.05277 2.309 0.021 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 749.25 on 745 degrees of freedom
Residual deviance: 742.32 on 742 degrees of freedom
AIC: 3211.3
Number of Fisher Scoring iterations: 4
| Resid. Df | Resid. Dev | Df | Deviance | Pr(>Chi) |
|---|---|---|---|---|
| 745 | 749.2497 | NA | NA | NA |
| 742 | 742.3237 | 3 | 6.926019 | 0.07429355 |
glm2 = glm(cover~elev, data = dat2, family = poisson)
summary(glm2)
anova(glm1,glm2,test="Chisq")Call:
glm(formula = cover ~ elev, family = poisson, data = dat2)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.0673 -0.8250 -0.3048 0.9991 2.1347
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.546e+00 5.135e-02 30.115 <2e-16 ***
elev -8.448e-06 5.471e-05 -0.154 0.877
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 749.25 on 745 degrees of freedom
Residual deviance: 749.23 on 744 degrees of freedom
AIC: 3214.2
Number of Fisher Scoring iterations: 4
| Resid. Df | Resid. Dev | Df | Deviance | Pr(>Chi) |
|---|---|---|---|---|
| 745 | 749.2497 | NA | NA | NA |
| 744 | 749.2259 | 1 | 0.02384893 | 0.8772699 |
glm4 = glm(cover~disturb*elev,data=dat2,family=poisson)
summary(glm4)Call:
glm(formula = cover ~ disturb * elev, family = poisson, data = dat2)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.4042 -0.7782 -0.2072 0.8090 2.0888
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.445e+00 1.396e-01 10.352 <2e-16 ***
disturbLT-SEL 2.546e-01 1.639e-01 1.554 0.1203
disturbSETTLE -3.702e-01 2.275e-01 -1.628 0.1036
disturbVIRGIN 9.283e-02 2.625e-01 0.354 0.7236
elev 3.540e-05 1.243e-04 0.285 0.7758
disturbLT-SEL:elev -2.788e-04 1.651e-04 -1.689 0.0912 .
disturbSETTLE:elev 7.319e-04 2.943e-04 2.487 0.0129 *
disturbVIRGIN:elev 2.278e-05 2.269e-04 0.100 0.9200
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 749.25 on 745 degrees of freedom
Residual deviance: 729.06 on 738 degrees of freedom
AIC: 3206
Number of Fisher Scoring iterations: 4
step(glm4)Call: glm(formula = cover ~ disturb * elev, family = poisson, data = dat2)
Coefficients:
(Intercept) disturbLT-SEL disturbSETTLE disturbVIRGIN
1.445e+00 2.546e-01 -3.702e-01 9.283e-02
elev disturbLT-SEL:elev disturbSETTLE:elev disturbVIRGIN:elev
3.540e-05 -2.788e-04 7.319e-04 2.278e-05
Degrees of Freedom: 745 Total (i.e. Null); 738 Residual
Null Deviance: 749.2
Residual Deviance: 729.1 AIC: 3206
The ANOVA contrasts suggest a significant difference in slope with elevation in the plots of prior settlement: disturbSETTLE:elev 0.0129 *; the step function shows that the interaction is necessary (large increase in AIC when the interaction is removed).
#?step#st=step(glm4)#st$anova